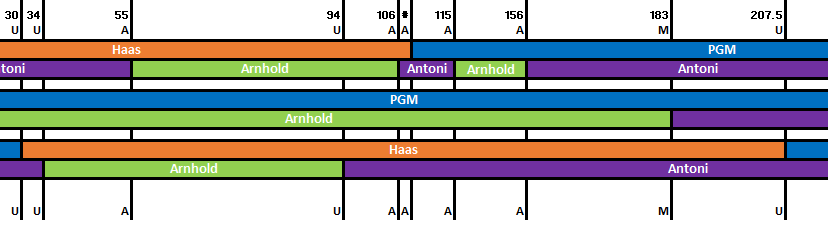
If you have the possibility to test three full siblings, then the next great thing you could do with your DNA results is to try out the Visual Phasing technique developed by Kathy Johnston. It allows you to map the segments of your chromosomes to your four grandparents (without having them or your parents tested) by comparing the recombination points of the three siblings. However, to figure out which of your grandparents a segment belongs to, you will still need additional cousin testing on several of your lines. Ideally, multiple 2nd cousins on either side of your family, but more distant cousins have proved to be very valuable as well.
I do not have full siblings, but luckily both my mother and father do. I have tested my mother and my maternal uncle a few years ago, and when I first read about Visual Phasing on Blaine Bettinger’s fantastic blog, I immediately knew what I was going to do next – get a DNA test for my maternal aunt, too!
In my earlier version of the post I was explaining how to do Visual Phasing manually, but I thought it was now time for an update, because I can no longer imagine doing Visual Phasing without Steven Fox’s marvellous automated Excel spreadsheet. It speeds up the process a lot! Therefore, after uploading autosomal raw data files of the three siblings to GEDmatch, you would need to join the Visual Phasing working group on Facebook – currently the only place, where Steven Fox’s Excel spreadsheet is available for download. However, you will only be able to use the automated spreadsheet, if you are already familiar with how the Visual Phasing method works and have a good understanding of genetic genealogy in general. So before we get started, let’s have a look at some basic information about genetic genealogy. (If you are already familiar with the basic concepts, please scroll down to the next part.)

Is it worthwhile to try this if one has just the kits of oneself and one living sibling plus a Lazarus kit of a deceased sibling to work with?
Thanks very much.
F.T.C.
Hello “Family Tree Climber”,
I wouldn’t use a Lazarus kit of a deceased sibling for VP. Instead, I would work with those kits you have used to create a Lazarus kit for your sibling directly in the spreadsheet. For example, if your sibling had children you can work with their data as Leanne Cooper did in her example https://leannecoopergenealogy.ca/2018/02/04/visual-phasing-with-two-siblings-and-a-niece-nephew/. I’ve also had success on some chromosomes using my maternal half sister and our phased kits for Visual phasing. If you have lots of close cousins, their data can be of huge help to determine crossover ownership!
Hi! I’ve been reading your blog for a long time
now and finally got the courage to go ahead and give you a shout out from
Dallas Texas! Just wanted to say keep up the fantastic work!
Thank you for your encouranging words! My blog is a rather slow development though, I’m focusing mostly on the German version at the moment.