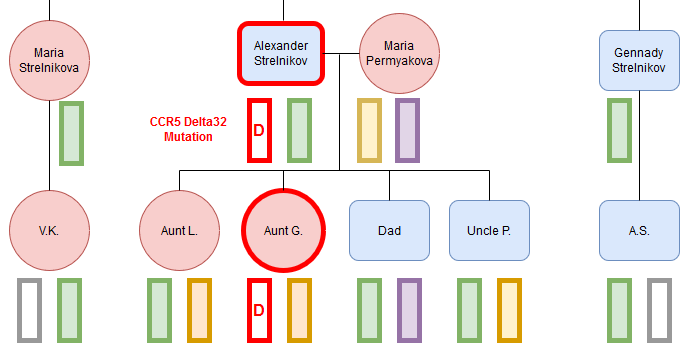
All companies that offer DNA testing for genealogical purposes also provide their customers with their DNA raw data files. Thus, depending on your interests and experience, this data can also be used for other things, such as learning a little more about your genetic predispositions. In addition to various third-party services that can analyze your data for a fee, with raw data you have the possibility to browse it all by yourself.